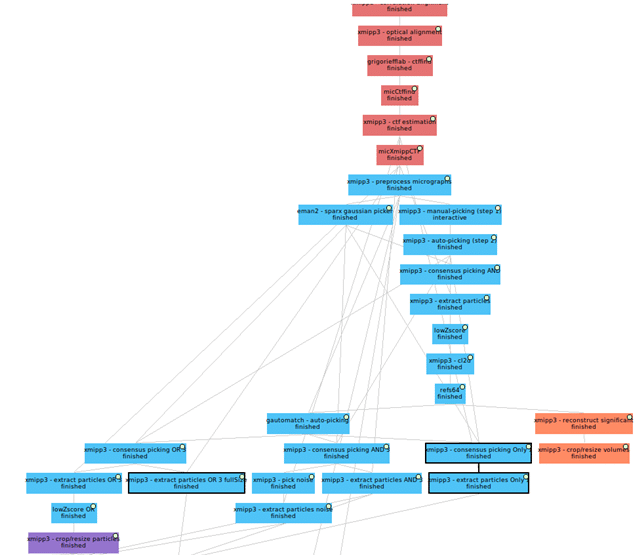
The CryoEM image processing use case in the EOSC Data Commons project aims to demonstrate how domain-specific scientific workflows can be seamlessly integrated into a federated cloud infrastructure. This use case focuses on deploying Scipion, a widely used open-source platform for CryoEM data analysis, on the cloud machines provided by the EOSC Data Commons.
Researchers can upload their raw CryoEM image data to the platform, where it becomes accessible for automated processing through predefined Scipion workflows. These workflows encompass the entire CryoEM analysis pipeline, including motion correction, contrast transfer function estimation, particle picking, and three-dimensional reconstruction. The orchestration and deployment of these analyses are managed using the platform’s underlying infrastructure, enabling efficient resource allocation and execution. This setup facilitates scalable and reproducible data analysis while also serving as a model for integrating advanced imaging tools into EOSC’s federated environment. The use case helps evaluate the platform’s ability to support complex, compute-intensive life science applications and provides insights into interoperability, metadata management, and automation requirements.